Welcome to DU!
The truly grassroots left-of-center political community where regular people, not algorithms, drive the discussions and set the standards.
Join the community:
Create a free account
Support DU (and get rid of ads!):
Become a Star Member
Latest Breaking News
Editorials & Other Articles
General Discussion
The DU Lounge
All Forums
Issue Forums
Culture Forums
Alliance Forums
Region Forums
Support Forums
Help & Search
Science
Related: About this forumThe Molecular Biology of Methicillin Resistant Staphylococcus Aureus Involves Sugar Chemistry.
The paper I'll discuss in this post is this one: Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity. (Peschel, Stehel et al Nature 563, pages705–709 (2018) )
A kind of mental block I've had over the years involves my understanding of the complex chemistry and biochemistry of sugars.
I've always found it hard, and tend to squirrel around it when it comes before me.
It's why I always love to read the introductory text of Gabius' The Sugar Code which I'm sure I've referenced before in this space.
To wit:
Teaching the biochemistry of carbohydrates is not simply an exercise in terminology. It has much more to offer than commonly touched upon in basic courses, if we deliberately pay attention to the far-reaching potential of sugars beyond energy metabolism and cell wall stability. In fact, then there is no reason why complex carbohydrates should shy at competition with nucleic acids and proteins for the top spot in high-density biocoding. On the contrary, sugars have ideal properties for this purpose, as will be concluded at the end of this chapter. In this sense, an obvious explanation why research in glycosciences (structural and functional glycomics and lectinomics) has lagged behind the fields of genomics and proteomics, also in the public eye, is 'that glycoconjugates are much more complex, variegated, and diffiailt to study than proteins and nucleic acids' [1]. What is a boon for decorating cell surfaces with a maximum number of molecular messages at the same time has been and still is a demanding challenge for analytical and synthetic chemistry…
H.J. Gabius ed. The Sugar Code, Wiley, 2009 pg 1.
That about sums it up. People don't think about sugars too much, because it's hard to do so...
(It's a great book by the way. I have to reference it all the time.)
One of the great - among many - challenges for future generations is antibiotic resistance. The extension of life span around the world is to a large measure the existence of antibiotics. My grandmother died at the age of 39 from a simple infection that could be cured today with a trip to the doctor, a prescription, and maybe 20 or 30 pills.
In order to address this rising crisis, it is useful to know how bacterial evolve to develop resistance to drugs, and, for that matter, natural antibodies.
This is why I found the paper cited above quite interesting.
From the introduction:
Novel prevention and treatment strategies against major antibiotic-resistant pathogens such as MRSA are urgently needed but are not within reach because some of the most critical virulence strategies of these pathogens are not understood8. The pathogenic potential of prominent healthcare-associated (HA)-MRSA and recently emerged livestock-associated (LA)-MRSA strains is thought to rely on particularly effective immune evasion strategies, whereas community-associated (CA)-MRSA strains often produce more aggressive toxins1,2. Most humans have high overall levels of antibodies against S. aureus as a consequence of preceding infections, but antibody titres differ strongly for specific antigens and are often not protective in immunocompromised patients, for reasons that are not clear3. A large percentage of human antibodies against S. aureus is directed against WTA5,9,10, which is largely invariant. However, some S. aureus lineages produce altered WTA, which modulates, for instance, phage susceptibility 7,11...
WTA is "Wall teichoic acid."
Teichoic acid, like nucleic "acid" is actually a copolymer.of a doubly phosphorylated acetylated aminosugar, galactosamine, and glycerol (or ribotol, an alcohol formed by reducing ribose.)
Here's a structure from Wikipedia:

The authors identified a protein called "TarP" that has close homology (27%) to a protein found in "normal" (not resistant) staphylococcus TarS. When they reinserted TarS into resistant strains, they found that they could restore susceptibility to antibiotics, as well as manipulate susceptibility to certain viruses.
I don't have a lot of time tonight, but here's some pictures and captions from the paper:
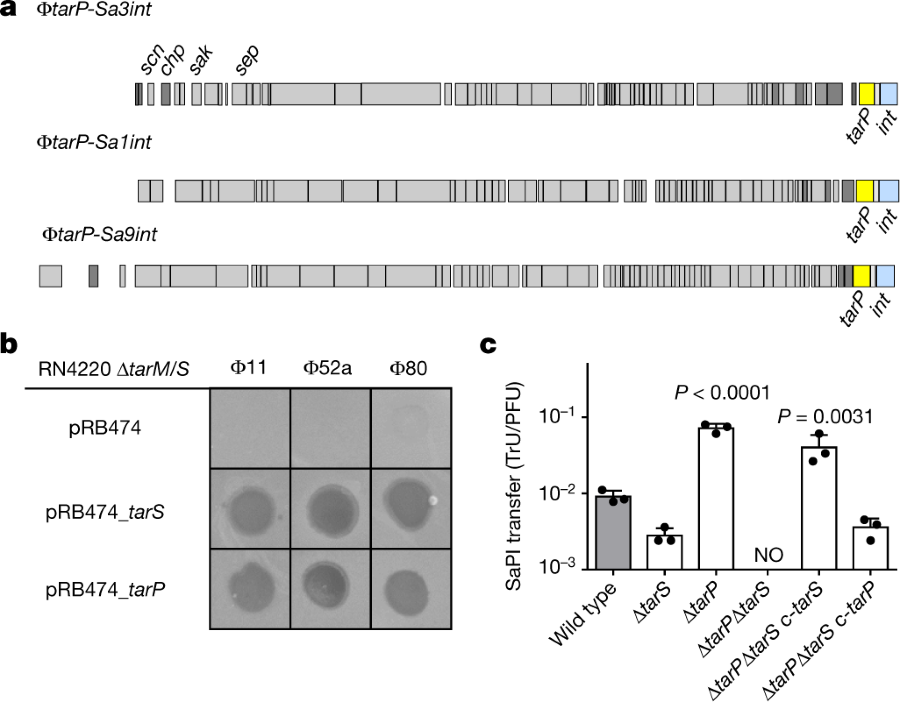
The caption:
a, TarP is encoded next to different integrase types (int gene) in prophages φtarP-Sa3int (with immune evasion cluster scn, chp, sak, sep), found in HA-MRSA, and φtarP-Sa1int and φtarP-Sa9int, identified in LA-MRSA. TarP variants in φtarP-Sa1int and φtarP-Sa9int differed from TarP in φtarP-Sa3int in one amino acid each (I8M and D296N, respectively). Both residues are distant from the catalytic centre. b, Complementation of S. aureus RN4420 ΔtarM/S with either tarS or tarP restores susceptibility to infection by WTA GlcNAc-binding siphophages, as indicated by plaque formation on bacterial lawns. Data shown are representative of three independent experiments. c, tarP expression reduces siphophage Φ11-mediated transfer of SaPIbov in N315. Values indicate the ratio of transduction units (TrU) to plaque-forming units (PFU) given as mean ± s.d. of three independent experiments. Statistical significances when compared to wild type were calculated by one-way ANOVA with Dunnett’s post-test (two-sided) and significant P values (P ≤ 0.05) are indicated. NO (none obtained) indicates no obtained transductants.
Some protein structures, teichoic acid structures, and some NMR's:
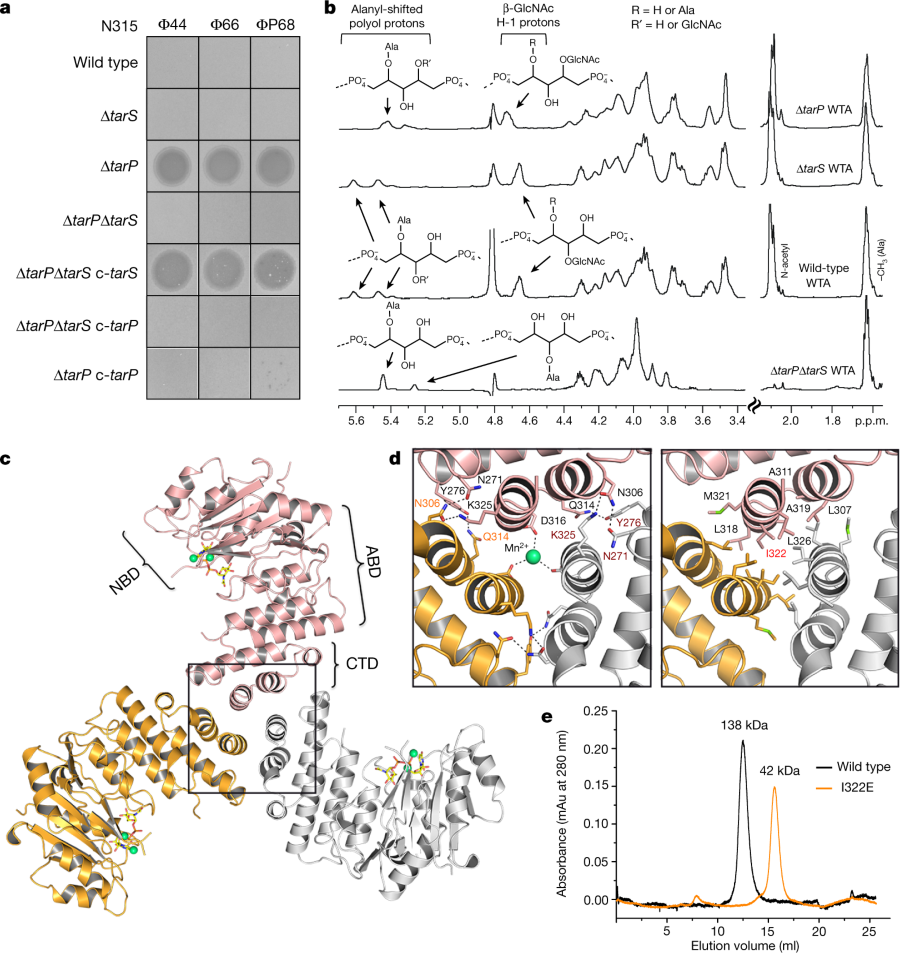
The caption:
a, Expression of tarP renders N315 resistant to podophages. Representative data from three independent experiments are shown. b, 1H NMR spectra reveal different ribitol hydroxyl glycosylation of N315 WTA by TarS (C4) or TarP (C3). The RboP units with attached GlcNAc are depicted above the corresponding proton resonances. Representative data from three experiments are shown. In-depth description of the structural motifs identified in the spectra is given in the Supplementary Information. c, Crystal structure of TarP homotrimer (pink, orange, grey) bound to UDP-GlcNAc (yellow) and two Mn2+ ions (lime green). The nucleotide-binding domain (NBD), acceptor-binding domain (ABD), and C-terminal trimerization domain (CTD) of the pink monomer are labelled. d, Views into the trimer interface (boxed in c). Left, polar interactions. Hydrogen bonds and salt bridges are shown as black dashed lines. The Mn2+ is 2.1 Å from each Asp316 carboxylate. Right, hydrophobic interactions, with the mutated residue Ile322 highlighted in red. e, Size-exclusion chromatography elution profiles. Based on calibration of the column, the TarP wild-type and I322E mutant proteins have estimated molecular weights of 138 kDa (n = 8) and 42 kDa (n = 3), respectively, in agreement with the calculated molecular weights of 120 kDa for a TarP trimer and 40 kDa for monomeric TarP.
A cartoon of interactions leading to the modified cell walss:
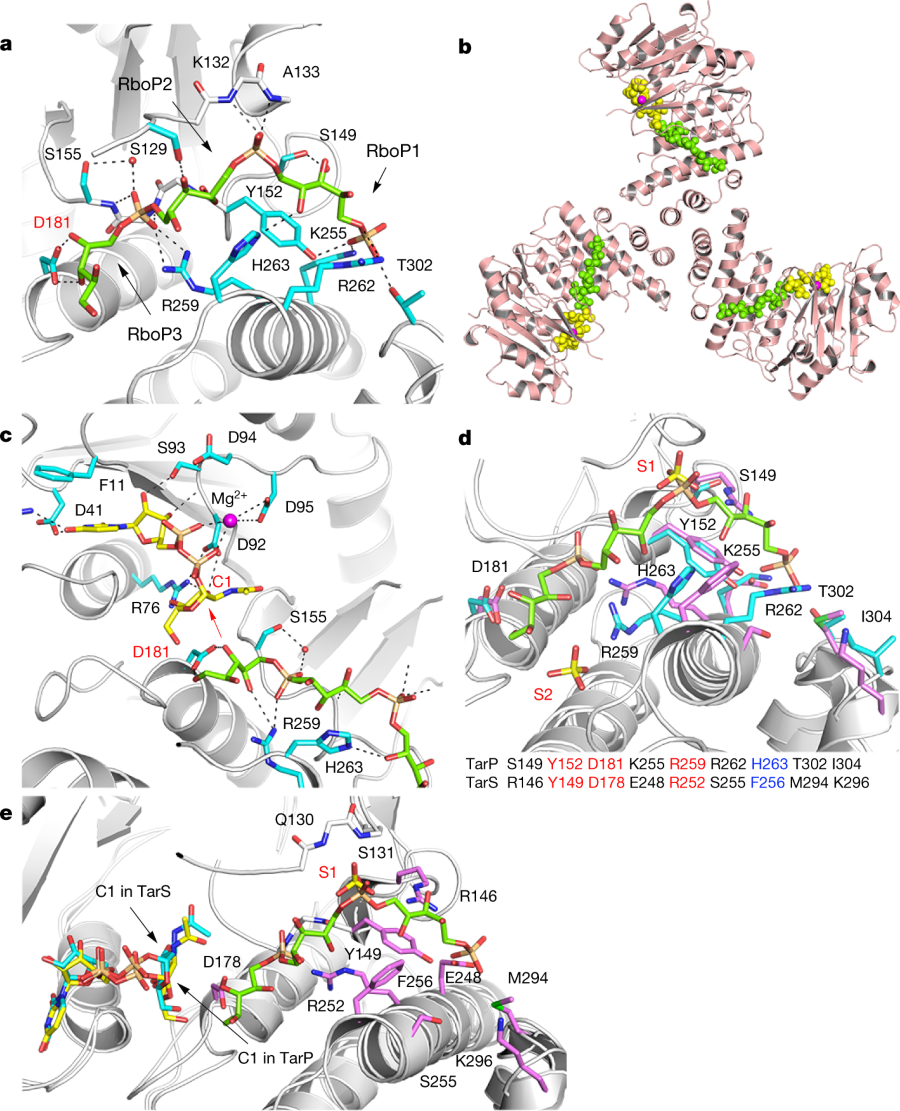
a, 3RboP binding site in the TarP–3RboP complex, with key amino acids shown (cyan). Asp181 is highlighted in red. The ribitol of 3RboP is coloured green and D-ribitol-5-phosphate units 1, 2 and 3 (RboP1, RboP2, and RboP3) are labelled. Hydrogen bonds and salt bridges are shown as black dashed lines. b, Ternary complex of TarP with UDP-GlcNAc and 3RboP. UDP-GlcNAc, Mg2+ and 3RboP are shown as full-atom models coloured yellow, magenta, and green, respectively. c, View into the active site of TarP. C1 of UDP-GlcNAc and Asp181 are highlighted with red labels. The arrow indicates how the C3-hydroxyl in RboP3 could nucleophilically attack GlcNAc C1. d, Comparison of the polyRboP-binding site of TarP with the corresponding region in TarS. Residues of TarP and 3RboP are coloured as in a. TarS residues are coloured violet and the two sulfates are labelled S1 and S2. Only residues of TarP are labelled, for clarity. Key TarP and TarS residues lining the polyRboP-binding site are shown at the bottom, with three identical (red) and one conserved amino acids (blue). e, Superposition of UDP-GlcNAc-bound TarS with the ternary TarP–UDP-GlcNAc–3RboP complex. UDP-GlcNAc and 3RboP bound to TarP are coloured as in b, whereas UDP-GlcNAc bound to TarS is coloured in cyan. Only the TarS residues are shown (coloured as in d), for clarity. The arrows indicate the C1 positions of UDP-GlcNAc in TarP and TarS.
Some immunology effects:
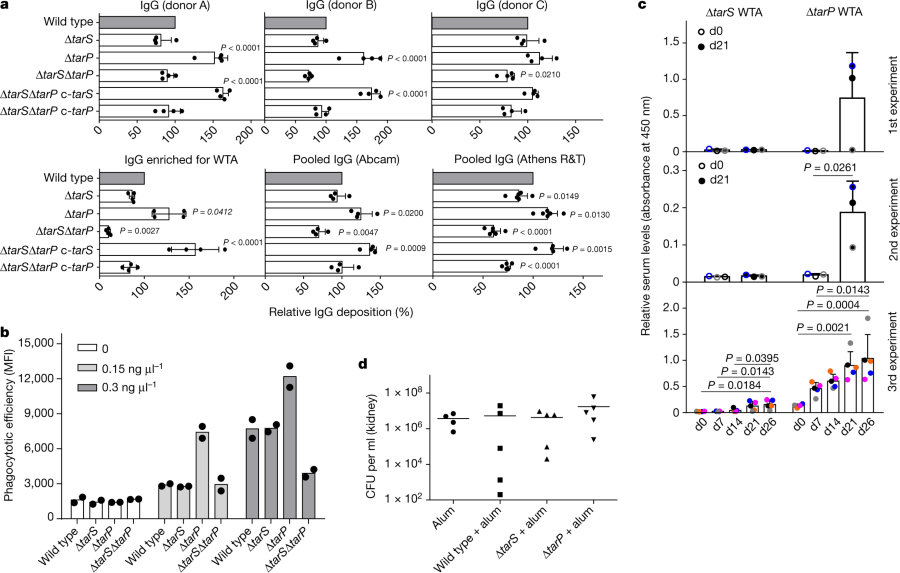
The caption:
a, TarP expression reduces deposition of IgG from human serum on N315 cells. The protein A gene spa was deleted in all strains. Top, human IgG isolated from three individual healthy donors (A, B, and C; n = 4); bottom, left, IgG from human serum enriched for RN4220 WTA binding (n = 4); middle and right, pooled human IgG from different suppliers (Abcam, n = 4; Athens R&T, n = 6). Results were normalized against wild type and shown as means with s.d. of n experiments. P values for comparison with wild type were calculated by one-way ANOVA with Dunnett’s post-test (two-sided), and P ≤ 0.05 was considered significant. Significant P values are displayed. b, TarP reduces neutrophil phagocytosis of N315 strains lacking protein A, opsonized with indicated concentrations of IgG enriched for WTA binding. Values are depicted as mean fluorescence intensity (MFI). Means of two dependent replicates of a representative experiment are shown. The other two representative experiments can be found in Extended Data Fig. 1g. c, TarP abrogates IgG response of mice towards WTA. For each experiment, WTA from N315 ΔtarP or ΔtarS was isolated independently. At least three mice per group were vaccinated and analysed for specific IgG at indicated time points after vaccination. Results are depicted as mean absorbance with s.d. Individual mice are indicated by colour. Increase in IgG levels was assessed by one-way ANOVA with Tukey’s post-test (two-sided). Significant differences (P ≤ 0.05) are indicated with corresponding P values. d, Vaccination with WTA does not protect mice against tarP-expressing N315, as shown for bacterial loads in kidney upon intravenous infection. No significant differences between groups of either five vaccinated mice or four mice for the alum control group (means indicated), calculated by one-way ANOVA, were observed.
Some concluding remarks:
Protection against S. aureus infections is urgently needed, in particular for hospitalized and immunocompromised patients2,4. Antibodies can in principle protect against S. aureus, but their titres and specificities vary largely among humans and they are often not protective in immunocompromised patients3, probably in particular against S. aureus clones that mask dominant epitopes, for instance using TarP. Unfortunately, all previous human vaccination attempts with protein or glycopolymer antigens have failed, for reasons that are unclear24. Our study identifies a new strategy used by pandemic MRSA clones to subvert antibody-mediated immunity, which should be considered in future vaccination approaches. S. aureus WTA with GlcNAc at RboP C3 has been reported as a type-336 antigen, but was not further explored25. We found that tarP is present in type-336 S. aureus (Extended Data Fig. 1f). However, TarP-modified WTA is a very poor antigen and vaccines directed against GlcNAc at WTA RboP C3 or C4 may fail against many of the pandemic MRSA clones. The structural characterization of TarP will instruct the development of specific TarP inhibitors that could become important in combination with anti-WTA vaccines or antibiotic therapies...
...TarP is a new and probably crucial component of the S. aureus virulence factor arsenal26,27, highlighting the important roles of adaptive immunity and its evasion in S. aureus infections.
...TarP is a new and probably crucial component of the S. aureus virulence factor arsenal26,27, highlighting the important roles of adaptive immunity and its evasion in S. aureus infections.
A note, which may or may not be all that easy to comprehend, from the front lines in confronting the growing antibiotic crisis with molecular biology.
Have a pleasant day tomorrow.
